Part:BBa_K3257070
SsrA(LAV)
This degradation tag has the amino acid sequence of AANDENYALAV. SsrA is a widely-used type of protein degradation tag that commonly consists of 11 amino acid residues and LAV stands for the last 3 amino acid residues of the tag. When fused after a protein, it will cause protein degradation to happen.
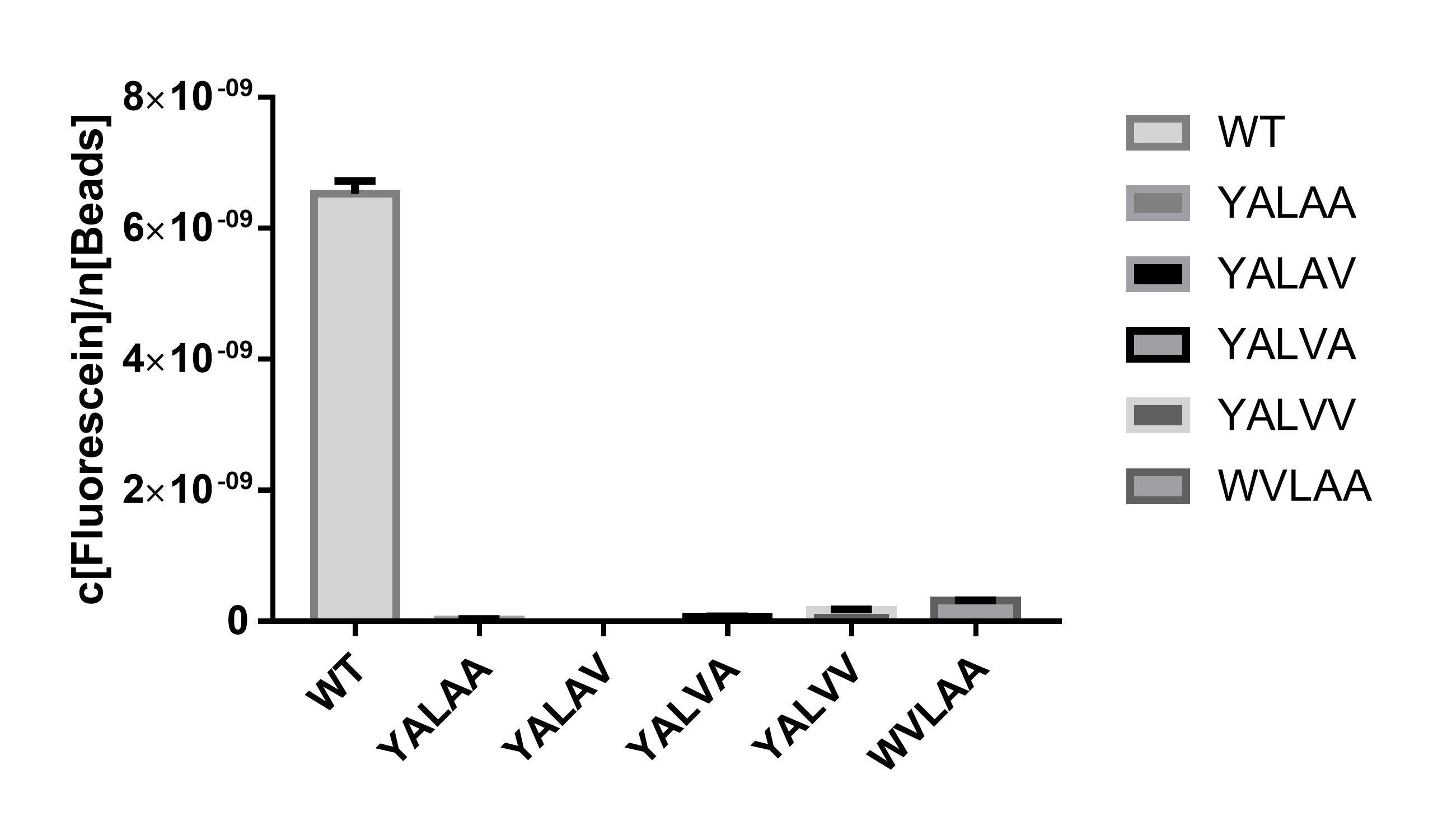
SsrA(LAA) (BBa_K3257069 https://parts.igem.org/Part:BBa_K3257069) is the wild-type SsrA degradation tag and the other 4 degradation tags are mutants of SsrA(LAA) which are expected to have nearly the same or lower degradation rate.
Using EGFP (BBa_E0040 https://parts.igem.org/Part:BBa_E0040) as a reporter protein, we have measured the fluorescent intensity of EGFP attached to different degradation tags at the steady-state (Figure 1), which indicates the degradation rate of each degradation tag when attached after a CDS of a protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
